Tumor Immune Evasion
John W. Vusich
2024-08-06
Tumor Immune Evasion of HPV+ Head and Neck Squamous Cell Carcinoma
Project Overview
This project analyzes the mechanisms of tumor immune evasion in HPV+ head and neck squamous cell carcinoma (HNSCC) by studying the epigenetic downregulation of MHC-I by PRC2. The study involves performing CUT&RUN on SCC90 HPV+ HNSCC cells treated and untreated with GSK126, an FDA-approved EZH2 inhibitor.
Methods
CUT&RUN
- Cell Line: SCC90 HPV+ HNSCC cells
- Treatment: Untreated (DMSO) and treated with GSK126
- Cell Number: 5 x 10^5 cells per target
- Replicates: 2 replicates per target
Library Preparation
- Kit: CUTANA ChIC/CUT&RUN kit (Epicypher, version 3)
- Purification: Qubit dsDNA HS kit
- Library Prep: CUTANA CUT&RUN Library Prep kit (Epicypher, version 1)
Sequencing
- Platform: NovaSeq 6000 (Illumina)
- Read Length: 2x50 bp
- Read Depth: 8-10 million reads per library
Data Processing
- Pipeline: nf-core/cutandrun v3.2.2
- Alignment: Bowtie2 v2.4.4 to GRCh38 and K12-MG1655
- Peak Calling: SEACR v1.3
- Consensus Peaks: Merged using bedtools v2.31.0
- Visualization: IGV desktop v2.16.1
Results
QC and Correlation Plots
Figure 1: Correlation Plot: 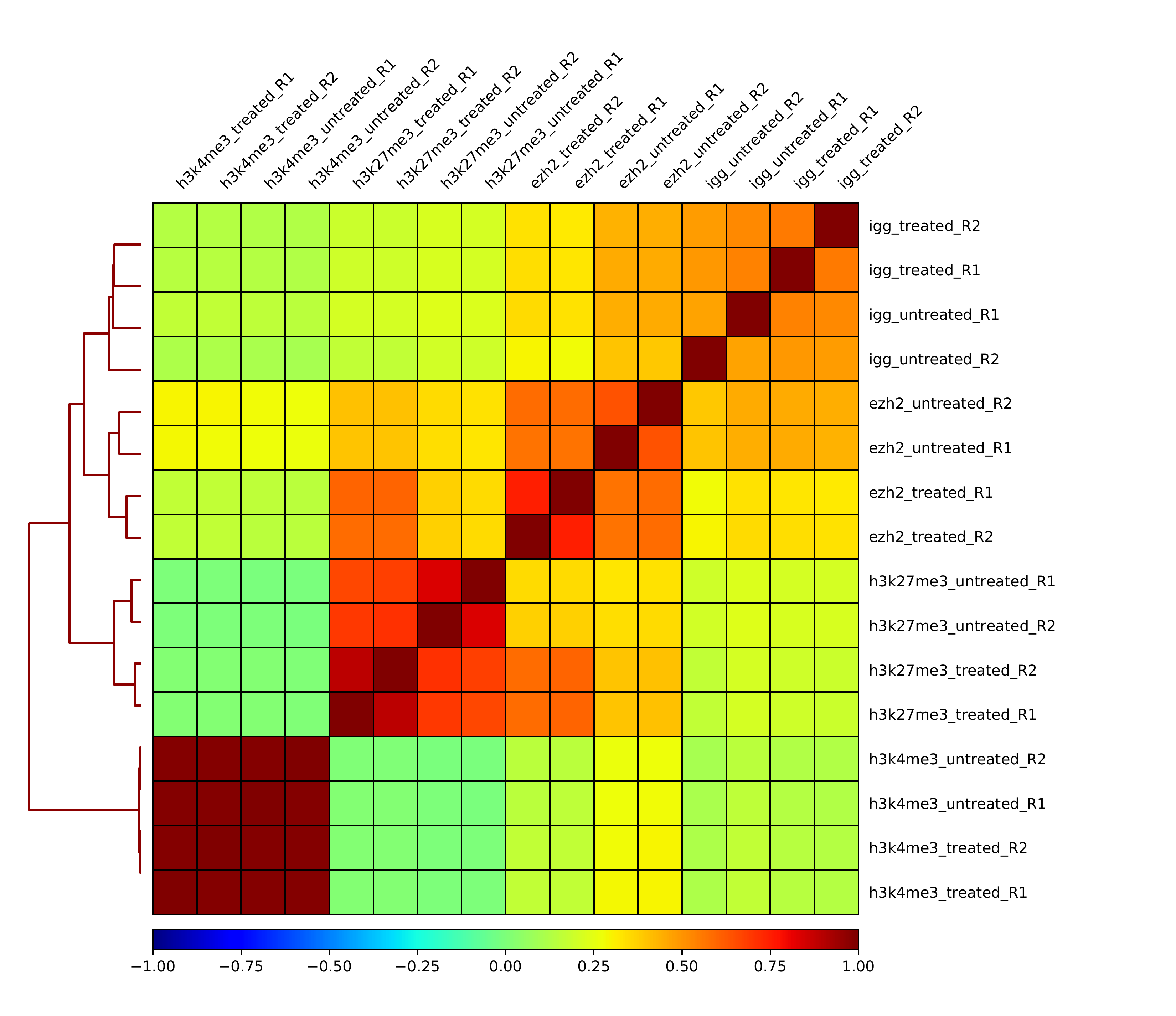
Peak Analysis
Figure 2: IGV Plot of HLA-B Locus: 
Differential Binding Analysis
Results from DiffBind and ChIPseeker analyses. Figure 4: MA
Plot: 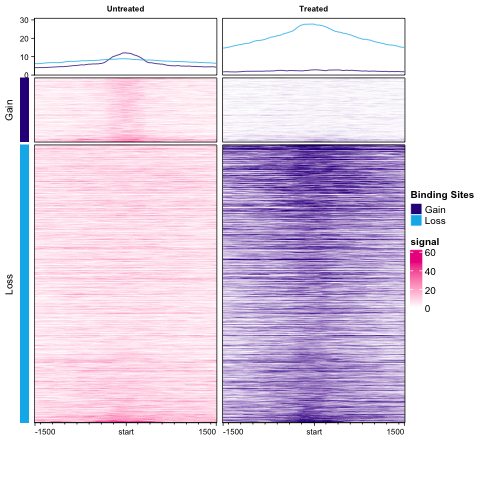
Figure 5: Volcano Plot: 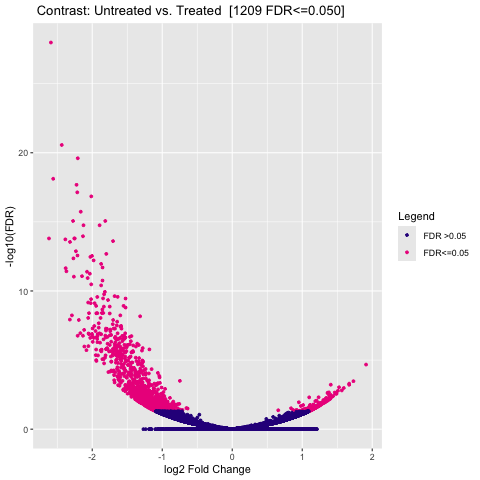
Figure 6: Heatmap: 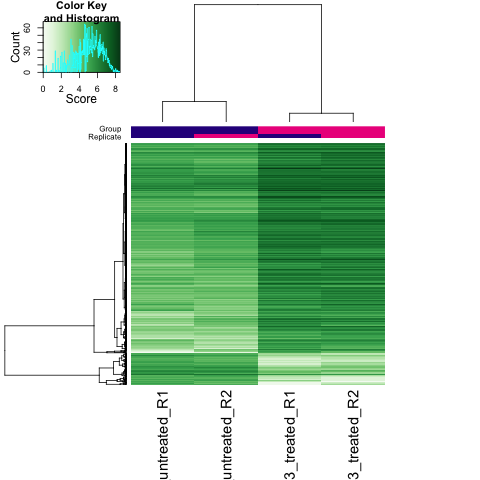
Figure 7: Venn Diagram: 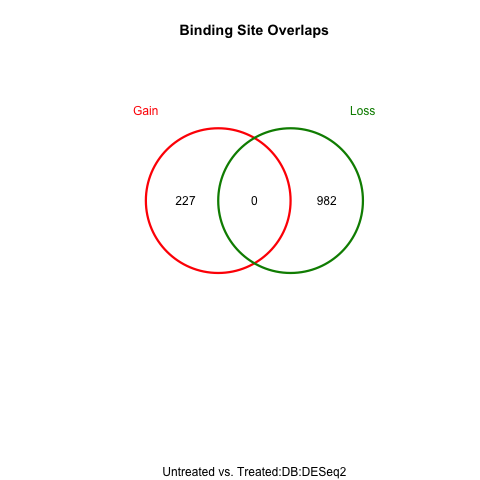
Figure 8: Profile Plot: 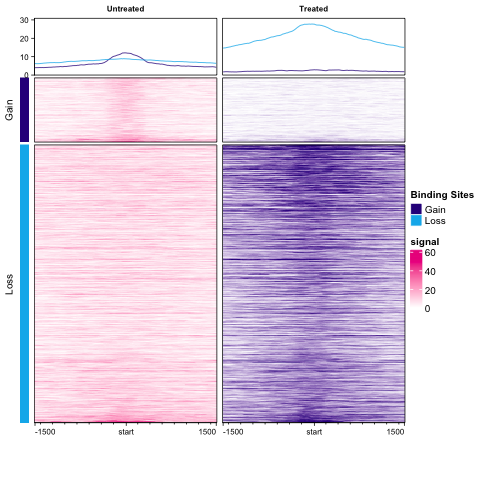
Conclusion
Our study found accumulation of H3K27me3 at the HLA-B and HLA-C loci, as well as several other MHC-I accessory/chaperone loci that was decreased following treatment with GSK126.
